16S v1-3 sequencing on MiSeq Supporting tagline
This is our protocol for sequencing 16S amplicons on the MiSeq. We use the Nextera XT barcodes as this is a quick and simple way of multiplexing 96 samples on each run. We have chosen v1-3 as it was a popular on the 454 but has only recently become suitable for the MiSeq with the introduction of the v3 reagent kits.
Primers
For our initial experiments targeting v1-3 we chose the popular primer combination 27F/519RF. Our primers also included ambigous bases to improve their coverage. This is a two-step PCR method, which has advantages in terms of scalability. In the first step you amplify your target region whilst adding Nextera XT overhang adapters. In the second step you add the barcoded adapters and perform additional amplification. The great thing about this method is you only need to order two primers to get started with 96 samples and you have the flexibilty to add further genes/targets down the road. Using the Nextera XT barcodes means you are less likely to have problems with poor % passing filter or % identified reads which can happen with homebrew constructs.
| Forward primer 5’-3’ | |
|---|---|
| Illumina overhang adapter | 27F |
| TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG | AGAGTTTGATYMTGGCTCAG |
| Forward primer |
|---|
| TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATYMTGGCTCAG |
| Reverse primer 5’-3’ | |
|---|---|
| Illumina overhang adapter | 519R |
| GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG | GWATTACCGCGGCKGCTG |
| Reverse primer |
|---|
| GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGWATTACCGCGGCKGCTG |
Our oligos are supplied by Alta Bioscience, Birmingham. Primers are usually lyophilised (freeze-dried) before shipping, so they need resuspending in PCR grade water. All necessary PCR equipment (e.g. pipettes, plastic ware) should be prewashed with 70% ethanol and left under UV for 15 minutes in a laminar flow hood. PCR reagents should also be mixed in a laminar flow hood.
First step PCR
| Reagent | Volume per reaction (µl) |
|---|---|
| 10x reaction buffer | 2.5 |
| MgCl2 50mM | 0.65 |
| dNTPs 10mM | 0.5 |
| F primer 10µM | 0.5 |
| R primer 10µM | 0.5 |
| Polymerase 5U/µl | 0.25 |
| PCR-grade water | 18.1 |
| DNA | 2 |
| Final volume | 25 |
| Step | Temp (°C) | Time (s) | Repeats |
|---|---|---|---|
| Pre-heat | 98 | 120 | |
| Denaturation | 98 | 30 | 20 |
| Annealing | 55 | 30 | 20 |
| Extension | 72 | 90 | 20 |
| Final extension | 72 | 300 | |
| Hold | 4 |
It is important to optimise the PCR conditions to suit your sample. Our samples were heavily contaminated with human DNA so in order to prevent excessive amplification of off-target human products, annealing temperature and MgCl2 concentration were optimised. This was done using a gradient PCR with a titration of MgCl2. A good starting point is an annealing temperature 5°C lower than the Tm of the primers and 1.5-4µl of MgCl2. We used BIOTAQ polymerase from Bioline with the recommended concentration of primers and dNTPs. All reactions were performed in triplicate and pooled to increase library diversity.
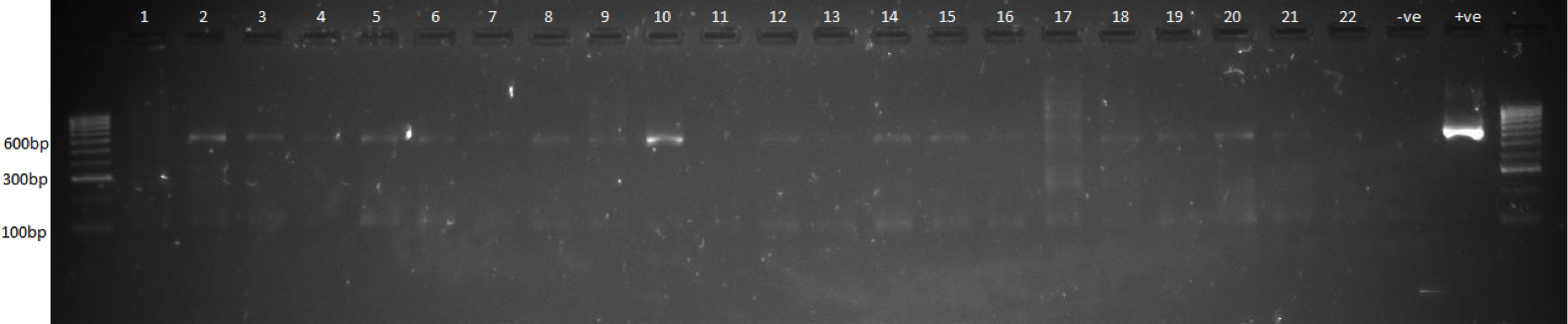
DNA gel showing the first step PCR amplicons before SPRI clean-up.
PCR products then need to be cleaned up using SPRI beads. The v1-3 amplicon is around 550bp in length so we used a ratio of 0.6x SPRI to clean-up the sample. This removes primers and any products below 400bp.
Second step PCR
As with the first-step the reaction conditions need to be optimised, however given that the overhang adapters should have an exact match for the barcoded adapters the optimum annealing temperature and MgCl2 concentration are likely to be higher. The Illumina barcode adapter plate is very useful at this step, as it holds the barcode tubes in position allowing you to pipette them with a multi-channel pipette. We do 10 cycles of PCR for the second step, after which the products (now 650bp) were run on a gel where they are all visible.
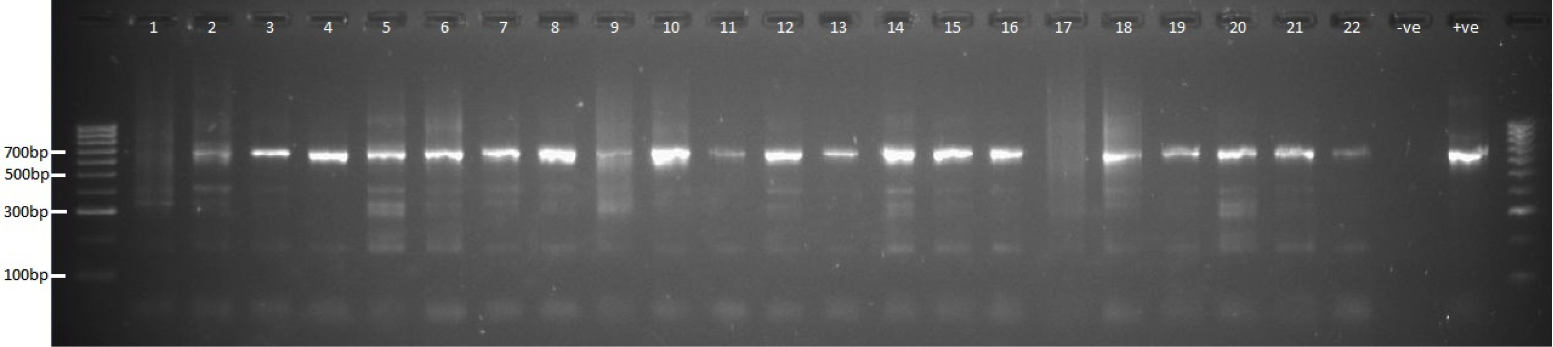
Products were cleaned-up again using 0.6x SPRI. A pool was made with equal volumes of each sample then quantified using SYBR Green qPCR with a KAPA library quantification kit.
KAPA quantification
The 96-well plate was made up in a laminar flow hood according to the manufacturers protocol and run on the Agilent Mx3005P instrument. Each reaction was set up in triplicate and treated collectively by the MxPro software.
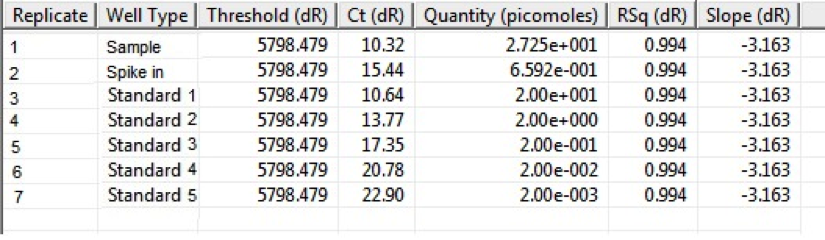
qPCR results displaying Ct values and initial quantity for samples and standards.
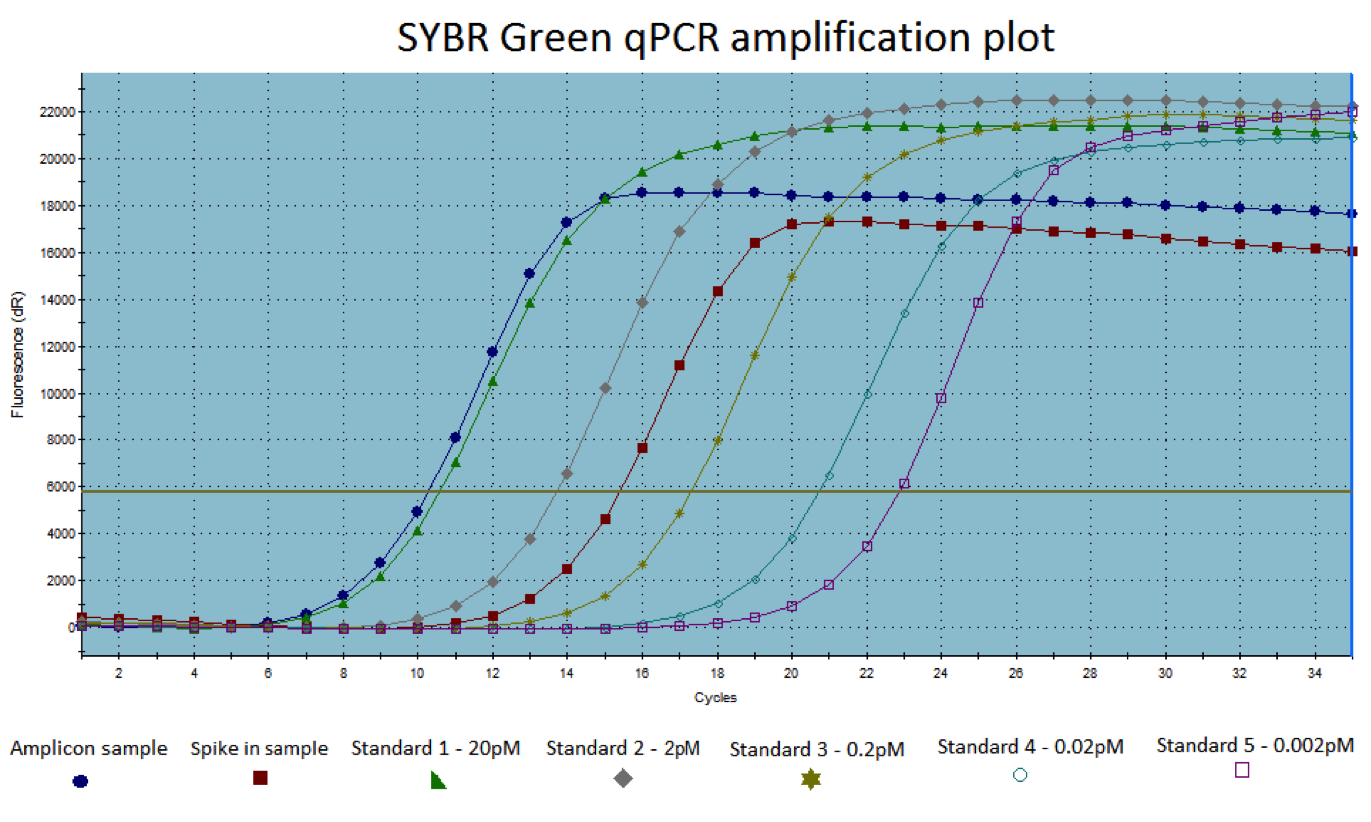
Amplification plots showing fluorescence by cycle for samples and standards.
MiSeq sequencing
Although recent improvements to RTA have overcome many of the problems assocaiated with sequencing low diversity libaries such as this, it is still necessary to use a spike-in. As the amplicon pool uses dual indexes it’s a good idea for your spike in to be dual indexed. We used a Nextera XT library taken from a previous plate, choosing an unused index combination. Using the quantification results, dilute both the amplicon pool and the spike in to 4nM with EB. Then add 8µl of the amplicon pool and 2µl of spike in (for 20% spike in) to an Eppendorf tube containing 10µl 0.2N freshly made NaOH, mixing up and down with a pipette. Leave this to incubate at room temperature for 5 minutes before adding 980µl of pre-chilled HT1. This gives you a 40pM denatured library, in order to target >20M reads load 40pM into a thawed MiSeq v3 reagent cartridge which should result in a density of >1000k/mm2. Run the MiSeq for 2x301 cycles which will take about 60 hours to complete.
Hello
blog comments powered by Disqus